Client
A company developing a healthcare platform hired Elinext to help it build a pneumonia diagnosis tool.
Challenge
The company has been developing a comprehensive healthcare platform. Treating pneumonia has been one of its focus areas due to COVID, and it wanted to build a pneumonia diagnosis tool for the platform.
The tool was destined to analyze lung X-ray images and identify signs of pneumonia using machine learning (ML), an artificial intelligence (AI) technique. The company didn’t have relevant in-house experts, so they reached out for help and found it with Elinext.
Solution
We began by looking for a neural network that would best analyze lung images and found four candidates: ResNet (50, 101, 152), VGG (16, 19), MobileNet, and Inception (V2, V3). After digging deeper into each of them, we chose InceptionV3 developed by Google Research Lab.
Once we chose our neural network, we moved on to designing the software architecture and training the algorithm.
Architecture
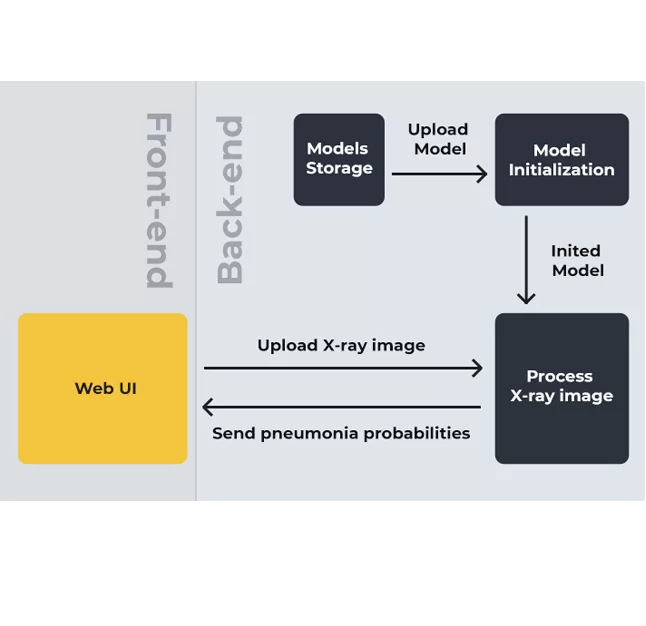
The software is based on web technology and can be integrated into other systems like desktop applications and mobile apps.
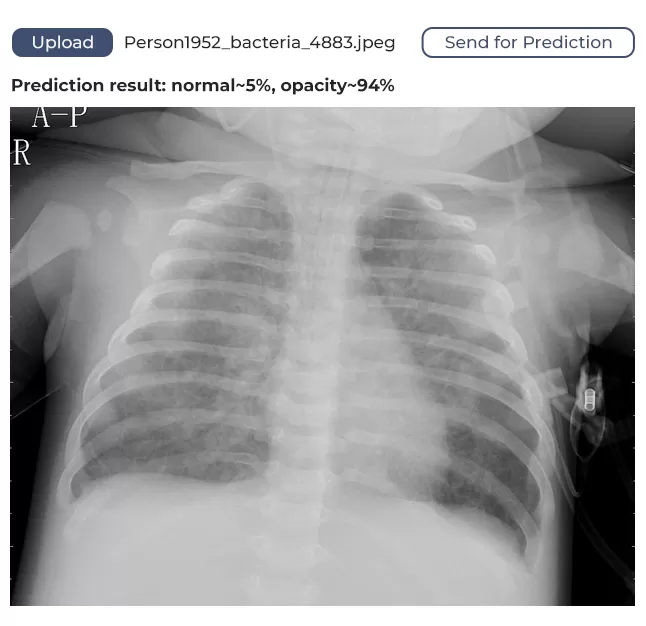
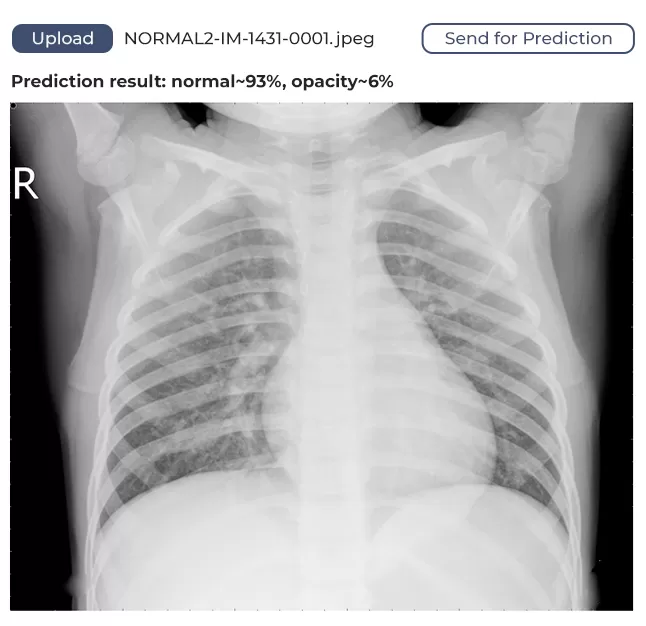
We used publicly available frameworks, libraries, and technologies to develop the software. To create a static HTML5 web page, we deployed a web server in a Docker container. On that page, a user can upload a lung image and get feedback. The image is sent for processing through the HTTP protocol.
Training
Training is the most challenging part of building ML algorithms. Your ability to source enough data, avoid errors and be consistent throughout the process can make or break the algorithm.
Manual training is often inconsistent. You may forget which steps you have taken and in which order, or occasionally delete logs. As a result, you won’t be able to accurately repeat a training session. Therefore, we automated the process from A to Z.
We needed to train complex models with huge datasets fast. To do that, we rented an Amazon Web Services (AWS) g3s.xlarge instance and used Deep Learning Base AMI (Ubuntu 18.10). The latter is a powerful machine boasting 16GB of RAM, a 4-core CPU, and an Nvidia Tesla M60 GPU. It was a perfect fit for the task.
Once we have chosen the technology, the training could begin. We built a clean Docker container to isolate the model from outer influences and downloaded a ton of lung images from Kaggle. To be able to work with the images, we subsampled them, narrowing them down to a relevant and consistent selection. The dataset and training environment were ready.
The training began. We faced a challenge in overtraining, whereby the model could memorize training images and as a result fail to accurately analyze new images in the future. Our solution was to slightly modify the images’ width, height, graininess, and some other parameters. We also launched Tensorboard to monitor training metrics.
At the final stages, we exported the model to an H5 file, a format commonly used across industries from healthcare to aerospace, for testing. We tested it manually and automatically, using preset scripts.
Accuracy
The model we’ve developed has a margin of confidence and uses binary identification. What does this mean? It means if the algorithm identifies 80% of lungs as unaffected, it will say the lungs are healthy. If the figure is below 80%, it will assume the lungs might be affected and require medical attention.